Abstract
The study of pharmacogenetics aids in the identification of variable therapeutic responses, identification of adverse reactions to drugs, and supports development and discovery. While advances in pharmacogenetic testing have been significant in recent years, The U.S. Food and Drug Administration (FDA) cautions against using this information solely as the basis to alter therapeutic treatments. In the case presented below, a patient with lung cancer was prescribed the standard dose of codeine for pain control. Pharmacogenetic testing confirmed that she was a normal metabolizer of CYP2D6 (CYP2D6 *1/*1). To manage the patient’s distress, fluoxetine, a high-affinity substrate of CYP2D6 was administered concomitantly. As the patient’s pain control became inadequate, the codeine dose was increased and co-analgesia with flecainide (another high-affinity substrate of CYP2D6) was initiated. The patient began hallucinating and became disoriented and confused. The concomitant administration of these drugs likely resulted in drug-induced phenoconversion. This case highlights the importance of correctly interpreting pharmacogenetic testing and of understanding drug-induced phenoconversion in the CYP450 metabolic pathway.
Introduction
Phenoconversion refers to a discordance between the clinically observed phenotype and the phenotype predicted from a patient’s genotype. For enzymatic systems involved in drug disposition, phenoconversion is especially important for patients with extensive enzyme activity such as ultra-rapid (UM), rapid or normal metabolizers (NM). In these patients, while their enzyme-related genotype predicts the use of regular drug doses (or even higher doses), drug biotransformation may be significantly impaired due to phenoconversion, leading to drug accumulation, increased systemic exposure, and toxicity. Phenoconversion generally results from nongenetic extrinsic factors such as food or drug intake, or changes in disease state [1].
This paper presents our interpretation of a case of phenoconversion where patient’s phenotype derived from genomic testing was likely altered by a multi-drug regimen. It tells a cautionary tale about interpreting pharmacogenomics results in isolation, and demonstrates how drugs metabolized through similar cytochrome P450 (CYP450) isoforms can compete to alter phenotypes. This illustrative case supports some of the concepts upheld by the FDA-issued warning letter to genetic testing labs for marketing genetic tests that claim to predict patients responses to specific medications [2]. It also exemplifies the importance of pharmacovigilance and of various ways to detect signals of drug related adverse events following drug entry to the market [3,4]. Various types of signal detection are clearly of importance especially under conditions of wide inter-subject variability in drug response due to pharmacogenetics and disease state.
The case example described here concentrates on interactions involving the CYP2D6 isoform. Before discussing details of the case, we will review some pharmacogenetic concepts and factors, such as concomitant medication that could lead to phenoconversion and modify a patient’s phenotype [1].
The term pharmacogenetics was first coined in 1959 and was used to describe phenotypic variation in drug metabolism [5]. In 1997, the term pharmacogenomics began to be used in conjunction with pharmacogenetics. While the terms pharmacogenetics and pharmacogenomics are sometimes used interchangeably in the literature, there are subtle differences between them [6,7]1.
Pharmacogenetic testing is an important tool that can be used to determine a patient’s genetic likelihood for an adverse drug event or drug toxicity, assess a patient’s risk for cancer and determine metabolic phenotype, all useful information that can influence therapeutic decision making [8]. As this is a rapidly evolving area of health research, there is still an incomplete understanding of the actionability of the test results. The quantity and variety of genes that each test analyzes can vary substantially, and the level of evidence required to determine whether a certain gene variant can affect phenotypes in a clinically significant manner is unclear [9].
The majority of pharmacogenetic tests are considered laboratory-developed and are regulated by the Clinical Laboratory Improvements Amendments [10]. Regulatory oversight from the FDA for pharmacogenetic tests was minimal until 2018, when 23andMe’s direct-to-consumer (DTC) products began to gain popularity. That year, the FDA declared permissible controlled marketing of 23andMe’s Personal Genome Service Pharmacogenetic Reports test, which analyzes 33 different gene variants, including CYP450 superfamily metabolic enzymes CYP2D6 and CYP2C19. With this policy, the FDA began exerting control over the information that the test reports provided. The FDA specifically stated that (a) the results from these DTC tests should not be used to alter medical treatments, and (b) the results should be verified by independent pharmacogenetics tests for accuracy before factoring into medical decisions [11]. In April 2019, the FDA cautioned genetic testing labs against providing medication use information to reduce the likelihood that consumers would change their own medication regimens in response to their test results [2]. Professional healthcare providers must interpret the results from these genetic reports, especially when multiple drugs are consumed by the patient.
Pharmacogenetic testing can add immense value to healthcare. Pre-emptive pharmacogenetic testing for CYP2D6 variants can be of great value to prevent serious drug reactions, like respiratory depression following opioid administration [12,13]. CYP2C19 genotype-guided warfarin dosing is available on www.warfarindosing.com and can be used to reduce the risk of bleeding [14]. CYP2C19 genotype-guided dosing can be used to determine clopidogrel dosing, or whether a switch to a different anti-platelet is required. In the field of oncology, genotyping a tumor type can give insight into which tyrosine kinase inhibitor will obtain optimum therapeutic success with chemotherapy [15].
More than 250 drugs include some pharmacogenetic information in their FDA labels. However, very few FDA labels actually recommend pharmacogenetic testing, or provide exact guidance for how therapy can be altered based on genetic information [16]. Various guidelines published by the Clinical Pharmacogenetics Implementation Consortium (CPIC), the Dutch Pharmacogenetics Working Group (DWPG), and the Canadian Pharmacogenetics Network for Drug Safety (CPNDS) help provide these missing links. With these guidelines, clinicians can better determine how to alter therapy based on pharmacogenetic information.
The Clinical Pharmacogenetics Implementation Consortium (CPIC) was established in 2009 to publish and maintain detailed clinical practice guidelines for drug-gene interactions. In 2014, CPIC published a clinical practice guideline for the CYP2D6 codeine drug-gene interactions. This guideline outlines how to interpret genetic test results for use in clinical practice when prescribing codeine, and how to determine a patient’s diplotype [17]. CPIC has published 23 guidelines to date for drug-gene interactions. Important pharmacokinetic genes, such as CYP2D6 and CYP2C19, along with pharmacodynamic genes, like CFTR and G6PD, have been included in these guidelines [18]. CPIC guidelines are considered a gold standard in the scientific community, and are endorsed by several professional societies [19].
The Dutch Pharmacogenetics Working Group (DPWG), established in 2005, is another group developing pharmacogenetics-based therapeutic dosing recommendations. Their goal is to integrate these recommendations into electronic drug monitoring systems to help both prescribers and pharmacists optimize medication use. DPWG publishes their work online on https://www.pharmgkb.org/, a well-respected knowledge resource for pharmacogenomics information. In 2018, DPWG updated its clinical practice guidelines regarding CYP2D6 and codeine use for patients with varying metabolizer statuses. This guideline outlines prescribing practices and provides recommended doses and more effective alternative medications [20].
The Canadian Pharmacogenomics Network for Drug Safety (CPNDS) has six published guidelines, including a codeine-specific clinical practice guidelines for CYP2D6 genotyping [12]. This guideline helps clinicians using genetic testing results both to determine patients who would not benefit from codeine therapy, and identify patients who are at risk for codeine toxicity.
Guidelines from these various authorities all recommend analyzing pharmacogenetic results in conjunction with a patient’s clinical history. Relying on a genotype guided phenotype may not be accurate if the patient’s metabolic phenotype is affected by their particular disease or by the drugs they are taking. This phenomenon is termed phenoconversion.
We fully endorse the position that pharmacogenetic results and clinical history must be reviewed in tandem, and we consider this comprehensive approach mandatory for optimal clinical interpretation of pharmacogenomic results. As such, we have developed pharmacogenomic models and implemented precision and predictive pharmacotherapy processes to improve outcomes for patients whose pharmacogenetic status is known and who are on a multi-drug regimen [21,22].
The concept of phenoconversion is less studied in pharmacogenomics, with only 124 PubMed results found while conducting searches on this term as of February 21, 2020. Drugs like quinidine that inhibit CYP2D6 can convert genotypic NMs, or possibly even UMs, to phenotypic poor metabolizers (PMs). There have been preliminary reports that inflammatory conditions and type 2 diabetes can influence the levels of CYP450 enzymes and can thus result in phenoconversion [23,24]. As the use of pharmacogenetics in healthcare increases, drug-induced phenoconversion will be a very important consideration. Here, we present a case where phenoconversion on CYP2D6 altered therapeutic treatment for a female patient.
1Pharmacogenetics applies to how variants in specific genes can affect the response to a single drug, while pharmacogenomics describes how the human genome can influence drug response.
Case Presentation
A 42-year-old female presented to her primary care physician (PCP) with shoulder pain. As the patient was not pre-emptively tested for CYP2D6, the clinician assumed a *1/*1 genotype. Codeine (15 mg QID) and acetaminophen (300 mg QID) were prescribed for pain management, and adequate pain control was achieved.
A few months later, an X-ray revealed a major mass in her right lung with metastasis to the collar bone. The patient was distraught and requested psychological and pharmacological support. She was prescribed fluoxetine 10 mg QD for one week, to be increased to 20 mg QD if necessary. Ten days later, she reported to her PCP that her pain had significantly increased, and the PCP consequently increased the codeine dose to 30 mg QID, and then to 60 mg QID.
One week later, she was admitted to the hospital for scheduled surgery to remove the mass in her lung. Flecainide 100 mg BID was added to her analgesic regimen due to poor pain control. Within two days, the patient presented with confusion and hallucinations. She could not recognize her family and exhibited aggressive behavior. A magnetic resonance imaging (MRI) was conducted to scan for potential metastasis to the brain, and results were negative. A clinical pharmacist recommended switching codeine and flecainide to morphine for pain control. Morphine was started at a low dose (10 mg/24 hours by IV infusion). Within two days, the patient’s psychological symptoms abated, and her pain was more effectively managed.
Discussion
Because 25% of all drugs are metabolized by CYP2D6, it is an important isoform in the hepatic CYP450 system [25,26]. CYP2D6 has been extensively studied and various clinically significant polymorphisms have been identified [20]. The *1/*1 variant is considered normal and an individual with this status is considered to be a “normal metabolizer” (NM). NMs exhibit what is considered the normal rate of metabolism. For these individuals, medications should work as intended [27,28]. In the case described herein, the patient was presumed to be a NM (which was confirmed later by genotype testing), but concomitant use of multiple CYP2D6 substrates with varying affinity resulted in severe drug interactions.
For an adult NM, the FDA recommends a codeine dose range of 15 mg to 60 mg every four hours as required, not to exceed 360 mg in a 24-hour window. Considerations, including a patient’s health status, opioid tolerance, type and severity of pain, and risk factors for addiction, should guide patient-specific codeine dosing [29]. CPIC and DPWG guidelines recommend use of FDA-label age- or weight- specific dosing for NMs. CPNDS also advises that codeine can be used per standard of care for those whose metabolism is considered ‘normal’ [12]. However, some research recommends to use caution when prescribing codeine to CYP2D6 NMs, as NMs could have additional risk factors for toxicity if receiving the maximal therapeutic doses of codeine [12,30,31].
For our patient, a NM, generally CYP2D6 substrates can be safely taken at normal doses. Thus, the initial prescription for codeine and acetaminophen was appropriate. Codeine is a weak affinity substrate for CYP2D6 and is metabolized to its active metabolite morphine, which is responsible for pain control. Acetaminophen is primarily metabolized by CYP1A2, so no interactions are expected from this combination (Figure 1), and the patient achieved adequate pain control.
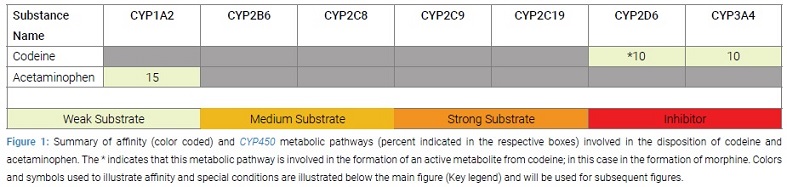
After her cancer diagnosis, fluoxetine was prescribed as an anti-depressant. Fluoxetine is a strong CYP2D6 substrate and is extensively metabolized by this enzyme (Figure 2).
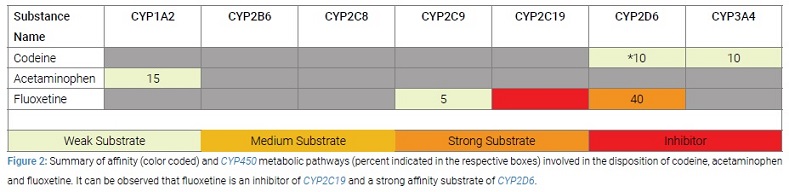
Fluoxetine also requires metabolization by CYP2C9 and inhibits CYP2C19. Since a substrate with stronger affinity for CYP2D6 was now vying for the active site on CYP2D6, the metabolism of codeine (a weak substrate) was diminished, inadequate morphine was converted, and the patient’s pain was not controlled. This context is an example of phenoconversion, where a NM phenotype was likely converted to a PM (poor metabolizer) phenotype when a stronger enzyme substrate was added to her regimen. Without recognizing and considering likely phenoconversion, the PCP increased the dose of codeine. Ideally, for a PM, the DPWG recommends prescribing morphine instead of codeine, as morphine is not metabolized by CYP2D6 [20].
The patient’s drug-gene problem was aggravated further when flecainide was added to her therapy as a co-analgesic. Like fluoxetine, flecainide is also a CYP2D6 substrate with strong affinity (Figure 3).
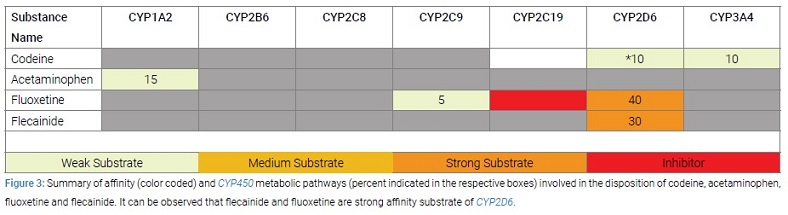
Now, two substrates with stronger affinity for CYP2D6 were competing for the receptor site that codeine requires for biotransformation. This completely blocked the transformation of codeine into morphine. Further, as flecainide is used at a higher dose than fluoxetine, plasma levels of fluoxetine likely spiked, resulting in the patient’s hallucinations and aggressive behavior.
MRI results came back normal and finally, a clinical pharmacist’s opinion was sought. Per the pharmacist’s recommendations, codeine and flecainide were stopped. Morphine was started at low dose, since morphine does not require CYP450-metabolism for conversion to an active form and is thus free of interaction-based complications (Figure 4).
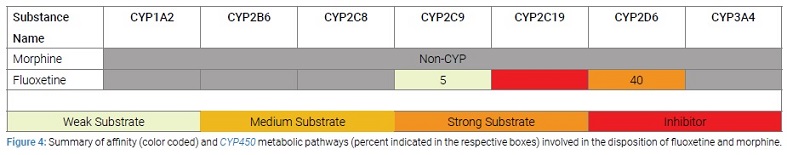
Adequate pain control was achieved.Fluoxetine was continued, and since there was no competition for CYP2D6, the adverse effects of fluoxetine disappeared.
Conclusions
Though the application of the rapidly evolving science pharmacogenetics is helpful in reducing trial-and-error prescribing, providers should use pharmacogenetic results in conjunction with a patient’s clinical history for safe prescribing practices. It is crucial that clinicians are informed and are diligent about the tricky phenomenon of phenoconversion. Despite the increase in the popularity of direct-to-consumer testing and the increase in the number of genes being tested, both providers and patients must be aware that phenoconversion should be a major consideration in medication reviews. Even a small number of concomitant medications can render genotype-guided phenotypes incorrect, resulting in over prescribing and dangerous prescribing cascades. Careful monitoring and report of adverse drug events by clinicians are important sources of information for appropriate use of pharmacogenetics correlated with pharmacovigilance.
Keywords
Phenoconversion; Cytochrome P450; Phenotype; Genotype; Pharmacogenetics; Pharmacogenomics
Cite this article
Deodhar M, Dow P, Al Rihani SB, Turgeon J, Michaud V. An illustrative case of phenoconversion due to multi-drug interactions. Clin Case Rep J. 2020;1(3):1–6.
Copyright
© 2020 Michaud V. This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International License (CC BY-4.0).




