Abstract
The benefits of opioid use in older adults to manage chronic non-cancer pain must outweigh the risks as these individuals are more susceptible to the side effects and drug interactions associated with opioids. Pharmacogenomic testing supports clinicians in selecting appropriate opioid therapy while minimizing these risks.
Abbreviations
CNCP: Chronic Non-Cancer Pain; PGx: Pharmacogenomics; CYP2D6: Cytochrome P450 2D6; CPIC: Clinical Pharmacogenetics Implementation Consortium.
Introduction
Given the widespread use and unfortunate misuse of opioids in the United States, the appropriate management of chronic non-cancer pain (CNCP) has been extensively debated in recent years. Of the 50 million adults with reported chronic pain, more than 60% are over the age of 65, and opioid prescribing is the highest in this population [1,2]. Older adults tend to have more persistent or severe pain and are more susceptible to the side effects and drug interactions associated with opioids than younger adults with similar CNCP [3]. Therefore, it is imperative that the benefits are optimized, and the risks are minimized when it comes to opioid use for CNCP in older adults.
The use of pharmacogenomics (PGx) testing effectively optimizes the benefits of certain drugs, such as antidepressants, clopidogrel, and warfarin [4]. PGx testing minimizes the risks associated with the opioid codeine. It could be used to balance the benefit-risk profile of not only codeine but all opioids that are metabolized by CYP2D6 [4]. This article describes a simulated patient case modeled after numerous encounters in our practice involving the clinical application of PGx for CNCP opioid optimization. We also provide examples of PGx-based recommendations to optimize treatment. This case serves as instructional guidance for clinicians only; medical decision-making will vary based on each individual patient case. Additionally, this case provides context about the clinical utility of PGx to guide opioid analgesic drug selection and demonstrates the impact of genetic variations and concomitant medications on drug interactions and phenoconversion.
Case Presentation
A 67-year-old Caucasian female presented with a past medical history of chronic, intractable lower back pain, hypertension, anxiety, and heart failure. Her physician prescribed several medications to manage her various conditions, including:
- Oxycodone tablets, 15 mg four times daily.
- Duloxetine capsules, 60 mg once daily.
- Lorazepam tablets, 1 mg twice daily as needed.
- Metoprolol succinate tablets, 50 mg once daily.
- Lisinopril tablets, 10 mg once daily.
At a follow-up appointment with her physician, she reported experiencing ongoing uncontrolled pain. Following multiple increases in her oxycodone dosage, from a total daily dose of 20 mg to 60 mg, she continued to experience inadequate pain control. When her physician sought assistance, the clinical pharmacist suggested that PGx testing would be helpful to assess the appropriateness of opioid therapy and guide medication changes. The physician then ordered a PGx test.
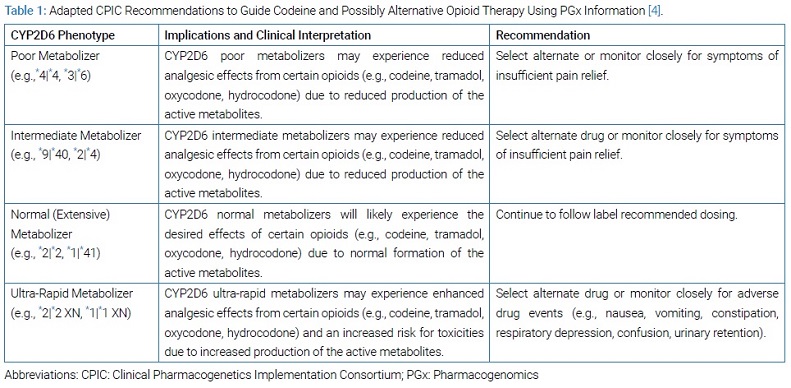
A DNA sample was collected by a buccal swab and analyzed by a genetics laboratory. The clinical pharmacist then interpreted the results. The patient was identified as an intermediate metabolizer for the cytochrome P450 2D6 (CYP2D6) drug-metabolizing enzyme, with a *9|*40 genotype (Table 1).
Considering the patient’s previous unsuccessful trials of non-opioid treatments (e.g., nonsteroidal anti-inflammatory drugs, skeletal muscle relaxants), the clinical pharmacist recommended that the physician consider an alternative opioid such as oxymorphone, hydromorphone, or morphine, which are not metabolized by the CYP2D6 drug-metabolizing enzyme [4]. After her physician changed the opioid from oxycodone to hydromorphone 2 mg four to six times per day as needed, the patient’s pain control improved significantly.
Importantly, the hydromorphone dose was established as if the patient was opioid-naive since the metabolic transformation of oxycodone into oxymorphone was impaired due to genetic polymorphisms, drug-drug interactions, and drug-drug-gene interactions. A direct dose calculation using morphine milligram equivalent dose algorithms was not applied, considering that the dose of oxycodone was increased up to 60 mg per day.
Discussion
Enzymes other than CYP2D6 are involved in the disposition of codeine, hydrocodone, tramadol, and oxycodone. For most of them, CYP3A4 and glucuronidation are the major metabolic pathways. Although CYP2D6 is only slightly involved in the total clearance of opioids, it is the major enzyme involved in activating these opioids for pain management. Consequently, CYP2D6 intermediate metabolizers have reduced enzyme activity that alters the CYP2D6-mediated activation of these opioids, reducing analgesic effects and increasing the risk of side effects [5,6]. (Table 2) allows users to view simultaneous multi-drug interactions when present.
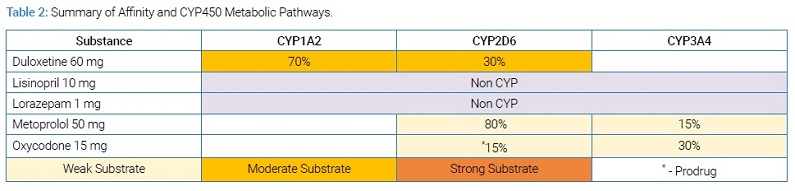
From this illustration, it can be seen that this patient was taking duloxetine, which is a known substrate with higher affinity than oxycodone for CYP2D6. Hence, when this drug is taken concomitantly with oxycodone, competitive inhibition of the CYP2D6 enzyme could occur [7]. Under these conditions, phenoconversion is observed (a disconnect between the genotype-predicted phenotype and likely phenotype observed clinically), and the patient’s CYP2D6 enzyme behaved like that of a poor metabolizer, which further reduced the activation of oxycodone into oxymorphone.
Several studies have demonstrated that single nucleotide polymorphisms or genetic variations, in the CYP2D6 gene can significantly affect the efficacy and safety of opioids [5,6]. There are numerous variant alleles, or different forms, of the CYP2D6 gene that an individual can possess that will result in decreased or increased enzyme function and, consequently, reduced or enhanced capacity to convert the aforementioned opioids to their active metabolites.
Studies have shown that individuals carrying copies of the CYP2D6 loss-of-function alleles, otherwise known as CYP2D6 poor metabolizers, exhibit lower concentrations of opioid active metabolites than normal metabolizers, resulting in reduced analgesia and poor pain control [5,6]. Another study in the post-surgical setting found that compared with CYP2D6 ultra-rapid metabolizers (individuals carrying multiple copies of the functional alleles), CYP2D6 poor metabolizers required a 33% higher dose of tramadol in the recovery room [8]. In an experimental study among a small group of healthy volunteers, researchers demonstrated a 1.5 to 6.0-fold increase of analgesic effect in CYP2D6 ultra-rapid metabolizers when compared to normal metabolizers; when normal metabolizers were compared to poor metabolizers, the latter experienced a 2 to 20-fold reduction of analgesic effects [5].
Yet another study showed that CYP2D6 ultra-rapid metabolizers had higher hospitalization rates than CYP2D6 normal metabolizers [9]. Due to increased rates of overdoses and fatalities among infants of breastfeeding mothers who were CYP2D6 ultra-rapid metabolizers and among children who were CYP2D6 ultra-rapid metabolizers receiving codeine, this opioid now carries a Black Box Warning in its prescribing information [10].
The Clinical Pharmacogenetics Implementation Consortium (CPIC) is an international organization facilitating the use of PGx for patient care. CPIC developed evidence-based guidelines for using PGx information to guide codeine and alternative opioid therapy [4]. These recommendations are summarized in (Table 1).
For CYP2D6 intermediate or poor metabolizers, CPIC recommends choosing an alternative opioid that is not metabolized by CYP2D6, such as hydromorphone or morphine [4]. The rationale is that the use of codeine and other opioids such as hydrocodone, tramadol, and oxycodone in patients with CYP2D6 intermediate or poor metabolizer status is associated with a high risk of reduced efficacy and poor pain control.
This case highlights the value of utilizing PGx information to enhance clinical decision-making and potentially improve therapeutic outcomes in patients with CNCP whose pain cannot be controlled with non-opioid therapy and require opioid therapy. Furthermore, it is important to note other factors should be considered in pain management (e.g., renal function, patient self-reported pain, concomitant medications).
Conclusion
In summary, CYP2D6 genetic variations and concomitant drug interactions leading to phenoconversion can significantly affect the therapeutic outcomes of patients receiving opioids. As illustrated by this simulated case, some patients cannot metabolize certain opioids to their active metabolites, which may result in poor pain control and diminished quality of life. The use of PGx information and drug-gene interaction data can help prescribers and other healthcare practitioners select the most precise opioid therapy for patients.
Acknowledgments
The authors want to thank Dana Filippoli for her comprehensive review and comments on the content of this paper.
Keywords
Opioids; Pharmacogenomics; Chronic pain; Non-cancer
Cite this article
Ballinghoff T, Bain KT, Matos A, Bardolia C, Turgeon J, Amin NS. Opioid response in an individual with altered cytochrome P450 2D6 activity: implications of a pharmacogenomics case. Clin Case Rep J. 2020;1(6):1–4.
Copyright
© 2020 Nishita Shah Amin. This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International License (CC BY-4.0).


